Root Parasitic Plants


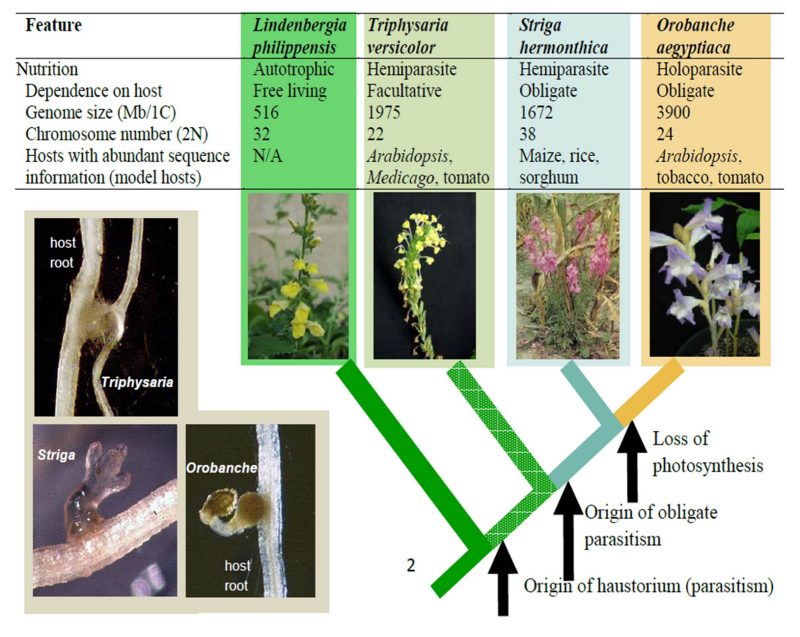
Parasitic plants derive part or all of their nutrients from another living plant. The parasites of the family Orobanchaceae attach to the roots of their hosts and draw nutrients from the vascular tissue. For example, the Phelipanche aegyptiaca parasite (left) is growing on the roots of a tomato host plant (right) in this picture. These parasites grow primarily underground, producing an an above-ground shoot only to flower.

The majority of species in the family Orobanchaceae are non-weedy, like the Conopholis americana (American cancer-root, shown at right) which grows on roots of oak and beach trees and is often seen along hiking trails around Virginia Tech. C. americana has no leaves and the only part of the plant that can be seen above ground is the pale flowering shoot.

Other parasitic plants in the family Orobanchaceae, however, cause agronomic devastation and severely limit food production in many parts of the world. The two obligate parasites Phelipanche aegyptiaca (right) and Striga hermonthica, along with other species in these genera, cause major agricultural damage. Weedy Orobanchaceae tend to thrive in warm, dry climates and grow primarily in Africa, the Mediterranean, the Middle East, and parts of Asia. For example, the species P. aegyptiaca grows on tomato, potato, tobacco, and other solanaceous crops in northern Africa, the Mediterranean, southern Europe, and South Asia. Striga hermonthica causes large reductions in corn, sorghum and millet crops in sub-Saharan Africa.
The PPGP takes a functional genomics approach to seek novel strategies that can be used for understanding and controlling the parasites, and for generating parasite-resistant crops.
The overall goal of the Parasitic Plant Genome Project (PPGP) is to carry out a comparative functional genomic analysis of parasitic plants in order to discover the genome-wide changes that led to the establishment of the parasitic lifestyle and the changes that resulted as a consequence of adoption of the parasitic life-style. Specific aims are to:
1. Identify genes associated with plant parasitism. Compare transcriptional profiles of haustorial tissues from selected parasite-host interactions to identify genes that are critical to enabling parasitism. These genes will be characterized in detail, with the most interesting ones studied by silencing and ectopic expression in parasites.
2. Characterize primary metabolism and specific nutrient transporters in parasite species that differ in their demands for host resources. Metabolite profiles will be developed for parasites that differ in host nutrient acquisition mechanisms. Genes that appear to be important for either nitrogen or sugar transport and metabolism will be characterized to understand their role in haustorial function.
Seeds of the root parasitic plants of the genus Orobanche germinate specifically in response to host-derived germination signals, which enables parasites to detect and attack preferred hosts. The best characterized class of germination stimulants is the strigolactones, although some species respond to sesquiterpene lactones such as dehydrocostus lactone. Despite great progress in characterizing the strigolactone signaling system in plants, the mechanism(s) by which parasite species detect specific compounds remains poorly understood. The goal of our project was to identify and characterize the genes responsible for stimulant specificity in O. cernua and O. cumana. These two species are closely related, but differ in host preference and stimulant specificity, with O. cernua responding to strigolactones and O. cumana responding to dehydrocostus lactone. We used a genetic approach based on O. cernua x O. cumana hybrids to associate germination response with genes. We found that these parasite species each have multiple copies of KAI2d genes, which function in strigolactone perception. For O. cernua, one of the KAI2d genes responds to strigolactone stimulants, but the dehydrocostus lactone receptor is not clear. Our data point to involvement of additional genes and yet greater levels of complexity regulating germination specificity in Orobanche.


