Research Projects
Research Overview
Major funded research projects include the following:

Dodder
The parasitic plant dodder (Cuscuta spp.) lives by attacking the stems of host species. Dodder makes vascular connections with its host, and takes all the resources it needs to grow and reproduce. During this process, both plants communicate in a molecular dialogue. We have discovered a bi-directional movement of a diverse range of RNA molecules, including mRNA and small RNA. However, we don’t know what roles these exchanged molecules are playing in the parasite-host interaction. To foster more research on this topic, we are optimizing methodologies for dodder research and developing new tools such as genetic transformation. This work will promote the study of dodder and facilitate research on plant interactions.
- See our Cuscuta Introduction page for general information and fun.
- See our Cuscuta Research Resources page for sequences, protocols, etc.
Collaborators: Michael Axtell, Penn State; Claude dePamphilis, Penn State
Funded by: NSF EDGE award IOS-1645027 (2017-2020) and NSF IOS-0843372 (2009-2014)
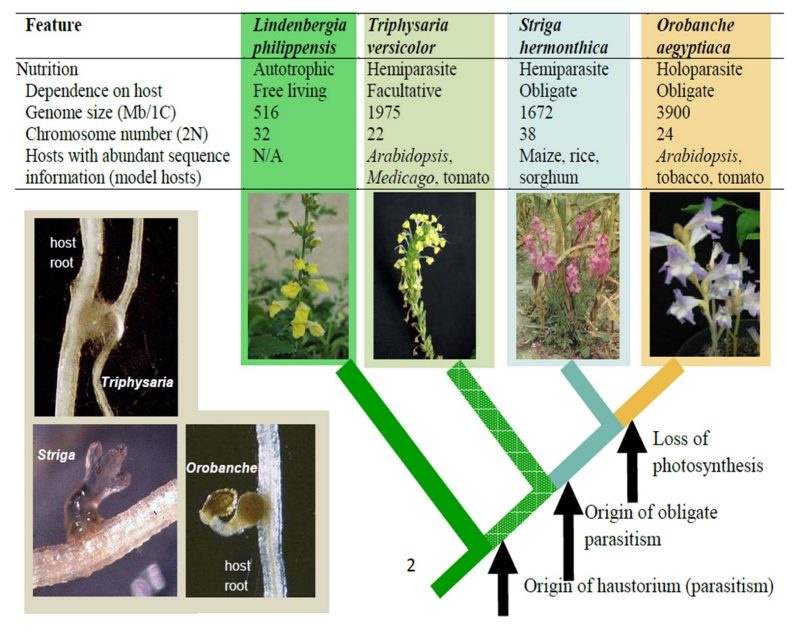
The Parasitic Plant Genome Project (PPGP)
The PPGP brings together researchers from around the US, along with international collaborators. We are studying the root-parasitic plant family Orobanchaceae, comparing key species that exhibit different levels of dependency on their hosts. Our project takes a functional genomics approach to investigate how parasitic plants have evolved in terms of development, metabolism, pathology, and genome evolution. Major goals include understanding the origin and evolution of the haustorium, and developing resources needed to produce parasite-resistant crops. The project hosts a transcriptome sequence database for species of the Orobanchaceae at the PPGP webpage: http://ppgp.huck.psu.edu/.
- See our Root Parasitic Plants page for more information
Collaborators: Claude dePamphilis, Penn State; Eva Collakova, Virginia Tech; Lenwood Heath, Virginia Tech; Michael Timko, University of Virginia; John Yoder, UC Davis
Funded by: NSF Plant Genome Research Program awards DBI-04-01748 (2007-2011) and IOS-1238057 (2013-2018).
Investigating the basis of seed germination specificity in Orobanche cernua and Orobanche cumana
Seeds of many species of parasitic plants in the family Orobanchaceae will remain dormant until they detect a host-derived germination stimulant. This unusual dormancy ensures that germination only occurs in the presence of an appropriate host. For most of these parasites, the germination stimulant is a strigolactone variant, but there are exceptions such as Orobanche cumana, which germinates in response to a non-strigolactone chemical. We are using these differences to understand the genetic mechanisms underlying germination stimulant specificity of parasitic plants.
Collaborators: Hanan Eizenberg, Danny Joel and Yaakov Tadmor, Newe Ya'ar Research Center, A.R.O. Israel; David Nelson, UC Riverside.
Funded by: US-Israel Binational Research Program (BARD) award no. US-4616-13 (2013-2017)
Epigenetics of weed response to stress
Weeds have been persecuted by humans since the dawn of agriculture, yet they persist and spread nevertheless. In recent years, weed species have shown remarkable capacity to evolve resistance to herbicides, thereby threatening the sustainability of modern agriculture. We hypothesize that the stress produced by weed control methods may in fact contribute to weed adaptation through epigenetic mechanisms (i.e., changes in DNA methylation, chromatin, and transposon activity that are not related to DNA sequences alone). The impact of such changes on broad-scale gene expression may allow weeds that survive initial control attempts to pass on adaptive patterns of gene expression that contribute an advantage to subsequent generations.
Collaborators: Shawn Askew, Jacob Barney, David Haak, and Liqing Zhang; Virginia Tech.
Funded by: USDA NIFA award 2017-67013-26593 (2017-2020)


